OBJECTIVE: Mathematical modeling plays an important role in understanding complex biomedical systems and the mechanisms underlying many pathologies, providing valid support to clinical studies. Mathematical models applied to the study of oncological and immunological diseases have reached high levels of sophistication and complexity, resulting in increasingly realistic and reliable descriptions in an effort to alleviate the human and economic costs involved. The workshop, therefore, aims to take stock of the state of the art of mathematics applied to oncology and immunology, to grasp emerging trends and strategies of interest not only to scholars in the field of mathematical and numerical modeling and bioinformatics, but also to doctors and biologists, and scholars of life/health sciences in general.
FUNDING
|
INdAM – Istituto Nazionale di |
Università degli Studi Mediterranea |
Dipartimento di Ingegneria Civile, |
Dipartimento di Ingegneria dell'Informazione, |
|
Consiglio Regionale della Calabria |
PROGRAM
INFORMATION
For info please contact A. Amoddeo or V. Bonanzinga at biomath-rc@unirc.it
Filippo CASTIGLIONE
- A virtual cohort study of SARS-CoV-2 infection and vaccination
Studies of the insurgence of immunity is at the core of both SARS-CoV-2 vaccine development and therapies. In this work I present an attempt to describe the insurgence (and the span) of immunity in COVID-19 at the population level using an in-silico model. We use a stochastic agent-based immune simulation platform to construct a virtual cohort of infected individuals with age-dependent varying degrees of immune competence. We use a parameter set to reproduce known inter-patient variability and general epidemiological statistics.
- In silico prediction of Tumor Associated Antigens’ immunogenicity
Computer simulations of anticancer immunotherapy can provide useful insights addressing the problem of protocol optimisation. I will describe the effect of varying dose and therapy administration on a cancer growth. A specific tumor type will be used in this example while the generality of the immune-cancer model will be highlighted.
Pasquale CIARLETTA
- Model and data fusion: physics-driven learning in cancer research
The key role of physical and mechanical interactions in cancer emerges from a very large variety of data sources and methods - from genomics to bioimaging, from proteomics to clinical records. Thus, learning physics-driven relational information is crucial to characterize its progression at different scales.
In this talk I will discuss how mathematical and computational tools allow for learning and better understanding of the mechano-biology of cancer, thanks to the integration of patient-specific data and physics-based models. I will present a few applications developed in the last decade in which the development of digital twins, empowered by ad-hoc learning tools, allows us to test new hypotheses, to assess the model predictions against biological and clinical data, and to aid decision-making in a clinical setting. (Funding from MUR - PRIN 2020, Progetto di Eccellenza 2023-2027 is gratefully acknowledged)
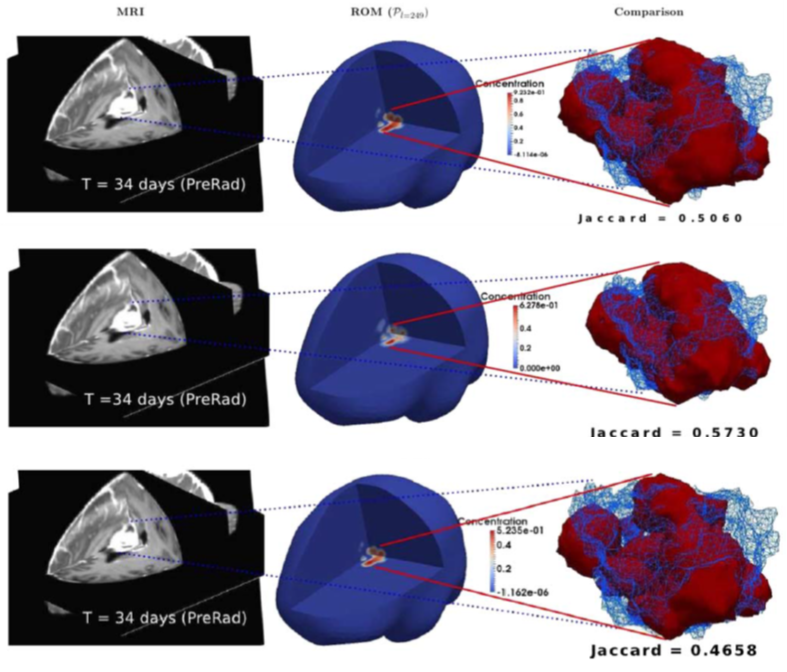
T cell therapy against cancer: A predictive diffuse-interface mathematical model informed by pre-clinical studies
T cell therapy has become a new therapeutic opportunity against solid cancers. Predicting T cell behaviour and efficacy would help therapy optimization and clinical implementation. In this work, we model responsiveness of mouse prostate adenocarcinoma to T cell-based therapies. The mathematical model is based on a Cahn-Hilliard diffuse interface description of the tumour, coupled with Keller-Segel type equations describing immune components dynamics. The model is fed by pre-clinical magnetic resonance imaging data describing anatomical features of prostate adenocarcinoma developed in the context of the Transgenic Adenocarcinoma of the Mouse Prostate model. We perform computational simulations based on the finite element method to describe tumor growth dynamics in relation to local T cells concentrations. We report that when we include in the model the possibility to activate tumor-associated vessels and by that increase the number of T cells within the tumor mass, the model predicts higher therapeutic effects (tumor regression) shortly after therapy administration. The simulated results are found in agreement with reported experimental data. Thus, this diffuse-interface mathematical model well predicts T cell behavior in vivo and represents a proof-of-concept for the role such predictive strategies may play in optimization of immunotherapy against cancer. (Funding from MUR - PRIN 2020, Progetto di Eccellenza 2023-2027 is gratefully acknowledged
Raluca EFTIMIE
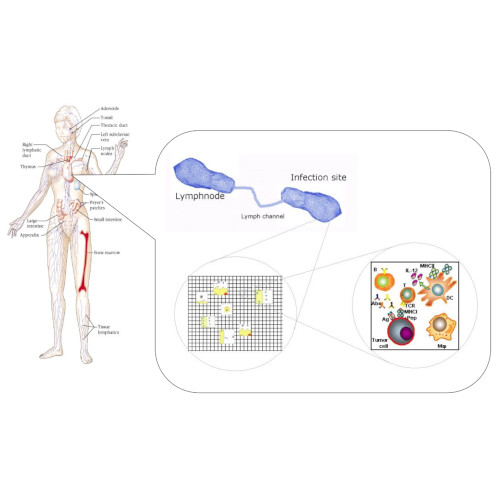
The development and evolution of cancers is associated with various immune responses, some pro-cancer and others anti-cancer, depending on the communication molecules (e.g., cytokines, chemokines, growth factors) present in the tumour microenvironment. The heterogeneity and plasticity of many of these immune responses, makes it difficult to fully understand the dynamics inside the tumour microenvironment.
In this talk I will focus on a particular immune cell population, the macrophages, which represent one of the first lines of defence against cancer. These cells form also one of the most abundant immune cell populations in some specific cancers (e.g., some breast cancers). While the roles of the two extreme macrophages phenotypes (the anti-tumour M1 cells and the pro-tumour M2 cells) are quite well known, experimental studies have also shown the existences of macrophages with mixed phenotypes, whose roles are still not fully understood.
Here, I will highlight a few single-scale and multi-scale mathematical modelling approaches aimed at shedding some light on the complex tumour roles of these immune cells with different phenotypes. In particular, computational approaches are used to propose new biological hypotheses on the potential role of macrophages with mixed phenotypes on delaying tumour relapse.
Viral infections are complex processes that occur across various time and spatial scales. The recent pandemics has brought into our attention the need for understanding such multi-scale interactions between viruses and the cells they infect, and further how these viruses might transmit between different individuals.
Here, we focus on both single-scale and multi-scale mathematical models for virus spread in the context of infectious diseases (e.g., COVID-19) as well as non-infectious diseases (e.g., cancer). While the modelling approaches are similar, the goal of the modelling is different: in the first case we try to understand and propose new hypotheses on the biological and epidemiological mechanisms that can stop the spread of the virus, while in the second case we try to understand and propose new hypotheses on the biological mechanisms that help the spread of the virus.
Francesco PAPPALARDO
- Beyond the Data: Computational Modeling as a Tool in Oncology and Immunology
- Simulating to Predict: Computational Models in Infectious Diseases
In Silico Medicine (the use of modelling and simulation in the support the clinical decision about individual patients) and in silico trials (the use of modelling and simulation in the assessment of new medical products) have the potential to revolutionize healthcare. The promises are enormous, but the risks associated to such radical departure from the traditional approaches is also considerable. Currently the journey of a new pharmacological product from the discovery of molecules until entering the market is very long and expensive. New interventions are first tested in vitro with cell cultures and then in vivo in animal models (usually two models are required to pass the approval of the regulatory bodies, the mouse being the most used one). Surprisingly, only 20% of the interventions that are successful in animals are then successful in patients during the most expensive phase of the assessment in clinical trials. The main target of biomedical world when dealing with the majority of pathologies is to understand its biological dynamics and to find a way to cure it. This heavily includes how cells and molecules of the immune system interacts each other to orchestrate a defense and how one can externally (and artificially) provide stimuli to eventually prepare human immune system in advance. Agent-based models (ABM for short) are computer models that attempt to capture the behaviour of individuals within an environment. Their wide usage in biomedicine environments is because they are more intuitive that mathematical or statistical models as they represent objects as individual things in the world. The most familiar examples to many people are The SIMs™ or SIMCity™ computer games in which people or other entities interact with each other and/or their environment. In biomedicine, ABM can be seen as in silico lab where one can capture the understanding of systems and, more fascinating, test “what if” scenarios. The lecture will provide a general introduction on what ABMs are and the main applications that involve their use in molecular and cellular modeling. In addition, a more detailed look at the biomedical applications is provided, with a special attention to immune system modelling, oncology and infectious diseases. Finally, a very brief state of the art towards regulatory context of in silico trials is presented
Luigi PREZIOSI
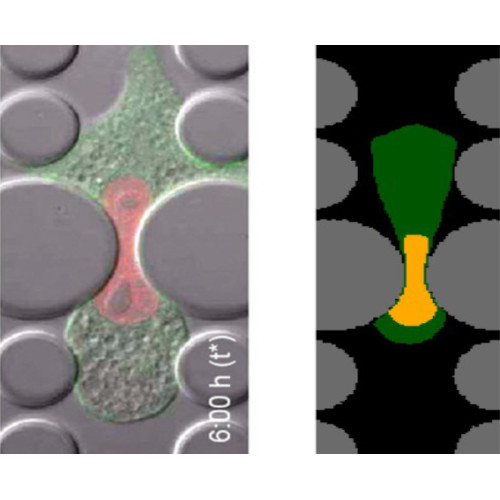
- Modelling Cell Reorientation under Stretch
The active response of cells to mechanical cues due to their interaction with the environment has been of increasing interest, since it is involved in many physiological phenomena, pathologies, and in tissue engineering. In particular, several experiments have shown that, if a substrate with overlying cells is cyclically stretched, they will reorient to reach a well-defined angle between their major axis and the main stretching direction. The aim of this talk will be to investigate the interplay between mechanics and cell organization. It will be shown that cells organise their internal structure to minimize an elastic energy that then drives this reorientation process. Viscoelastic effects will then be included to explain the dependence of the appearance of the phenomenon as a function of oscillation frequency.
- Multi-level mathematical models for cell migration in dense fibrous environment
Cell-extracellular matrix interaction and the mechanical properties of the cell nucleus have been demonstrated to play a fundamental role in cell movement across fibrous networks, micro-channels and therefore the microstructure of porous scaffolds. So, their study is important to understand both motion and growth in confined environments and also the spread of cancer metastases. This talk will merge the results of some continuum mechanics models and individual cell-based models that take into account of cell adhesion mechanics and nucleus mechanical properties to finally deduce a macroscopic model able describe the motion and growth in dense fibrous environments.
Carmelo TUSCANO
Radiation therapy, alongside surgery and chemotherapy, has historically constituted one of the three pillars of solid tumor treatment in humans. While the role of the immune system in tumor prevention has been recognized for over a century, recent advances in development of immune checkpoint inhibitors drugs have led to concrete clinical applications. Randomized trials have demonstrated statistically significant survival benefits across various tumor histotypes. An active area of research — both basic and clinical — focuses on exploring the correlations between immunotherapy drugs and radiation therapy to maximize therapeutic synergy.
Ionizing radiation has the ability to generate neo-antigens in response to the damage it induces at the cellular and subcellular levels. These neo-antigens stimulate mechanisms known as Danger-Associated Molecular Patterns (DAMPs), which, in turn, trigger an amplification of immunological signaling. This process potentially reinforces the disinhibitory effect on CD8+ T lymphocytes mediated by immune checkpoint inhibitors.
The aim of this talk is to present the radiobiological basis of synergies between radiotherapy and immunotherapy underlining that, despite progress, several questions remain unanswered. Optimal timing for the use of either immunotherapy or radiotherapy, the most suitable dose fractionation scheme to optimize synergy, and the definition of appropriate treatment volumes are areas of ongoing investigation. We present literature data that reveal promising clinical outcomes associated with the immuno-radiotherapy approach. The conclusion is a clinical case excerpted from the activities of the Department of Radiotherapy at the Grande Ospedale Metropolitano of Reggio Calabria.
Organizers:
- Antonino AMODDEOUniversità Mediterranea di Reggio Calabria
- Vittoria BONANZINGAUniversità Mediterranea di Reggio Calabria
Direttore Unità di Ricerca INDAM per Reggio Calabria - Vincenzo GIACOBBEUniversità Mediterranea di Reggio Calabria
- Pasquale GIOVINEUniversità Mediterranea di Reggio Calabria
- Luigi PREZIOSIPolitecnico di Torino
Technical staff:
- eng. Maurizio CAMPOLODICEAM
- Dr. Giandomenico POSILLIPODIIES
Orientation and Mentoring Service:
-
Dr. Simona VitaleHead of the Sector Orientation and Mentoring
-
Dr. Angela ViglianisiAdministrative Official of the Sector Orientation and Mentoring
-
Dr. Vania CalabreseCollaborator of the Sector Orientation and Mentoring
-
Dr. Wilma SessaCollaborator of the Sector Orientation and Mentoring